Protocols
DynaCLR
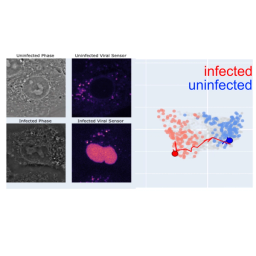
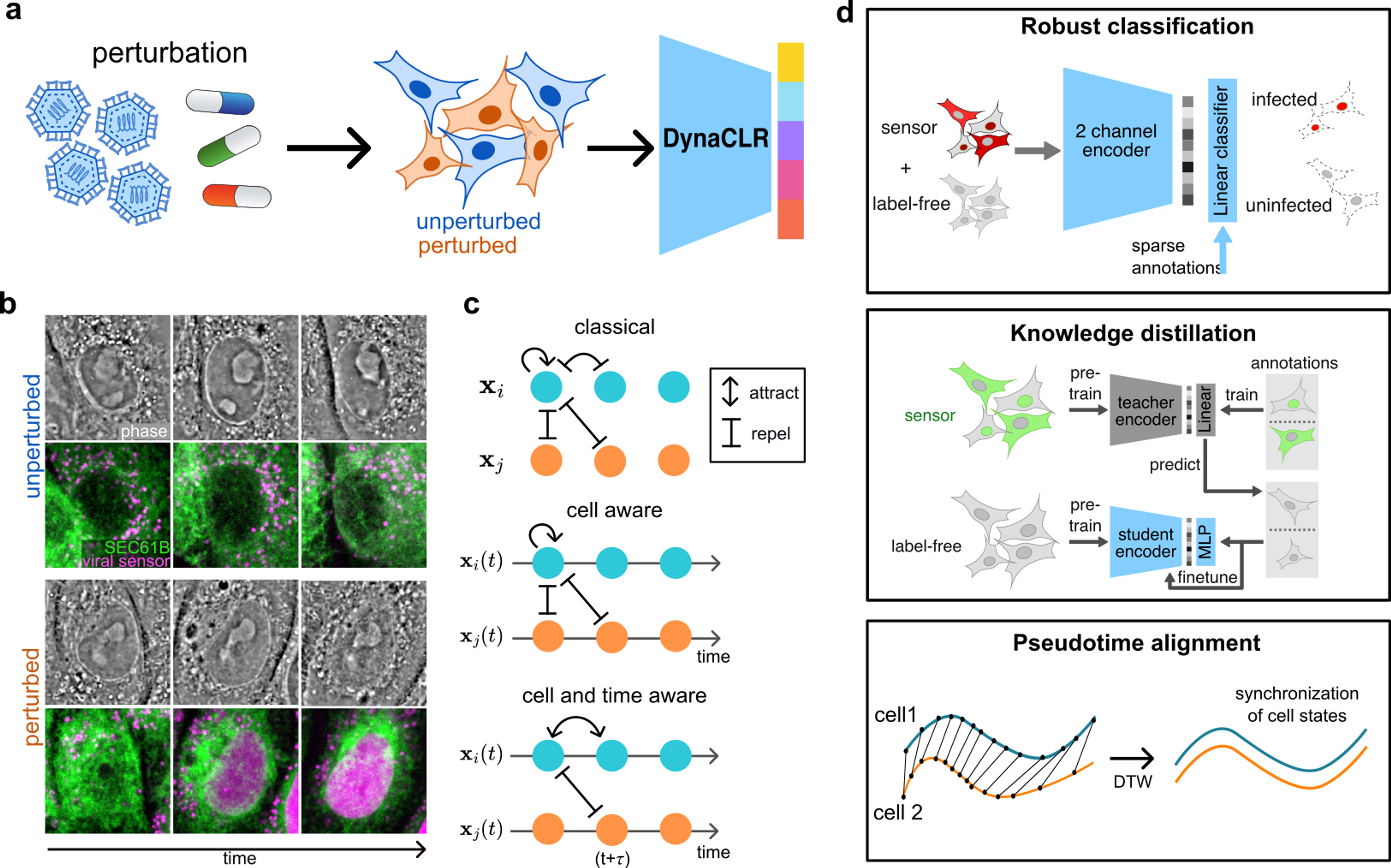
We have developed self-supervised deep learning method to analyze the cell and organelle dynamics in response to perturbations, DynaCLR (Dynamic cell state analysis using Contrastive Learning of Representations).
Demos
The DynaCLR demos with setup and running instructions can be found here